Troubleshooting a behavioral optogenetics experiment
Challenge
During his PhD, Nick Edwards (Potato's CEO) , ran an optogenetic experiment stimulating nucleus accumbens terminals in VTA. Based on some circuitry he'd discovered, he expected robust reward-related behavior. Instead, the results were murky: inconsistent trends, high variability, and no clear signal. Nick spent over a week trying to analyze the data - cleaning files, running statistics, revisiting the literature, and comparing histological data. But he never reached a definitive conclusion and the results remained unpublished.
Solution
Years later, we gave the exact same dataset and a relevant paper studying the same circuitry to Tater. (Yang et al. 2018) From a single prompt, Tater:
- Parsed and cleaned the raw CSV (142×10 table, messy headers, mixed data types) into a tidy 136-row dataset; standardized categories; and derived key behavioral metrics such as stimulated-side preference.
- Conducted statistical tests at both row- and animal-levels: one-sample tests, between-group t-tests (ChR2 vs YFP), matched pairs, and effect size calculations.
- Visualized the results with publication-ready plots (distributions, per-animal trajectories, matched-pair differences).
- Generated a review of Yang et al., 2018, mapping where Nick's experiment diverged from the published design.
- Identified factors contributing to the failed experiment through active troubleshooting.
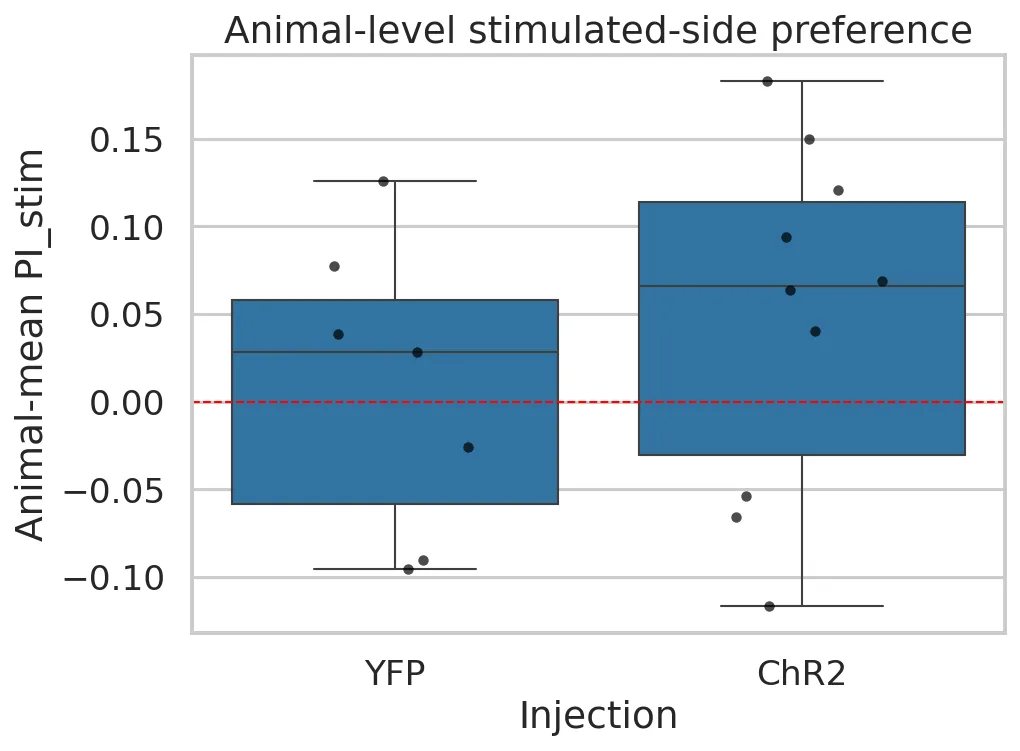
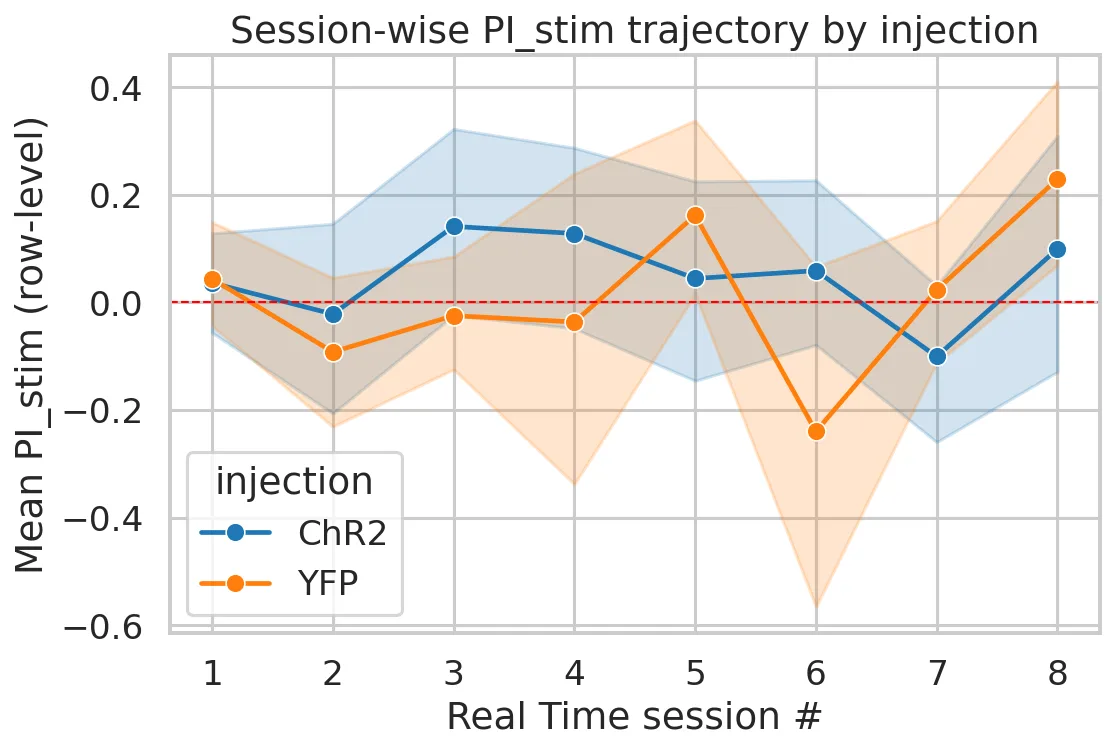
Statistical Results
- Group effect: Experimental mice showed a small positive effect compared to control mice, but differences were not statistically significant (all p > 0.15). Effect size was modest (d ≈ 0.43) with high uncertainty.
- Individual variability: A single “responder” ChR2 mouse consistently preferred the stimulated side (7/8 sessions), while most animals clustered near zero.
- Deliverables: A reproducible data analysis notebook, figures illustrating null results and variability, and a literature comparison bridging Nick's dataset with Yang et al., 2018.
Troubleshooting
Tater also produced feedback regarding possible reasons the data did not yield results matching Yang et al. Reviewing the feedback, Nick thought the injection placement was a likely culprit, supported by the extensive validation in Yang regarding injection targeting.
-
Comparison to Yang 2018: The findings did not reproduce the strong place
preference reported for NAcLat→VTA stimulation. Tater identified likely reasons:
- Mixed or unverified targeting of medial vs lateral NAc projections.
- Stimulation parameters (frequency, pulse width, irradiance) may not have matched effective ranges.
- Absence of pharmacological manipulation (e.g., GABA_B blockade) that Yang showed could unmask reward in NAcMed fibers.
-
Troubleshooting suggestions:
- Verify virus injection and optical fiber placements histologically (NAcLat vs NAcMed, medial vs lateral VTA).
- Add within-animal light-on/off blocks or sham sessions to reduce variance.
- Pair stimulation with a GABAB antagonist to unmask potential aversive effects.
Impact
For Nick, this experiment was one of the most frustrating episodes of his PhD: weeks of effort, inconclusive results, and no publication. With Tater, the same dataset was analyzed and contextualized in a few minutes with a simple prompt. The agent not only showed why the results were ambiguous but also generated a roadmap for what to try next.
This experience captures the motivation behind Potato: Nick's own time as a scientist was wasted trying to diagnose experiments that didn't work.An AI collaborator could have delivered clear, literature-grounded answers within minutes. Today, with Tater, researchers don't just get a statistical result - they get diagnosis, context, and actionable next steps. The result is faster iteration from data → insight → next experiment, turning what once felt like a dead-end into a learning tool and a foundation for better science.
Ready to try it yourself? Get started for free with our open access plan.
Citations
- Yang W, Carrillo-Reid L, Bando Y, Peterka DS, Yuste R. Simultaneous two-photon imaging and two-photon optogenetics of cortical circuits in three dimensions. Elife. 2018 Feb 7;7:e32671. https://doi.org/10.7554/eLife.32671