Automating AAV qPCR on a liquid handler
Challenge
Quantifying recombinant AAV vector genomes for gene therapy product release is a critical assay that demands careful design and strict regulatory alignment. Scientists not only choose appropriate primers, probes, and pretreatments, but also ensure the workflow complies with regulatory expectations and ICH Q2(R2) validation requirements. This process typically requires cross-disciplinary expertise: biologists to design and build the assay run by hand, automation engineers to get that protocol running reproducibly on a liquid handler, and QA specialists to confirm results, controls, and reporting standards. Translating this process into a robust, automated protocol can take weeks if not months of effort, slowing down preclinical programs and risking reproducibility.
Solution
We prompted Tater:
“Make an automated protocol for an Opentrons OT-2 instrument to perform qPCR quantification of AAV… Search the literature for appropriate methods, compare across papers… write a detailed protocol including what data analysis needs to be done. Then translate it to an automation script.”
From this single request, Tater delivered an end-to-end package:
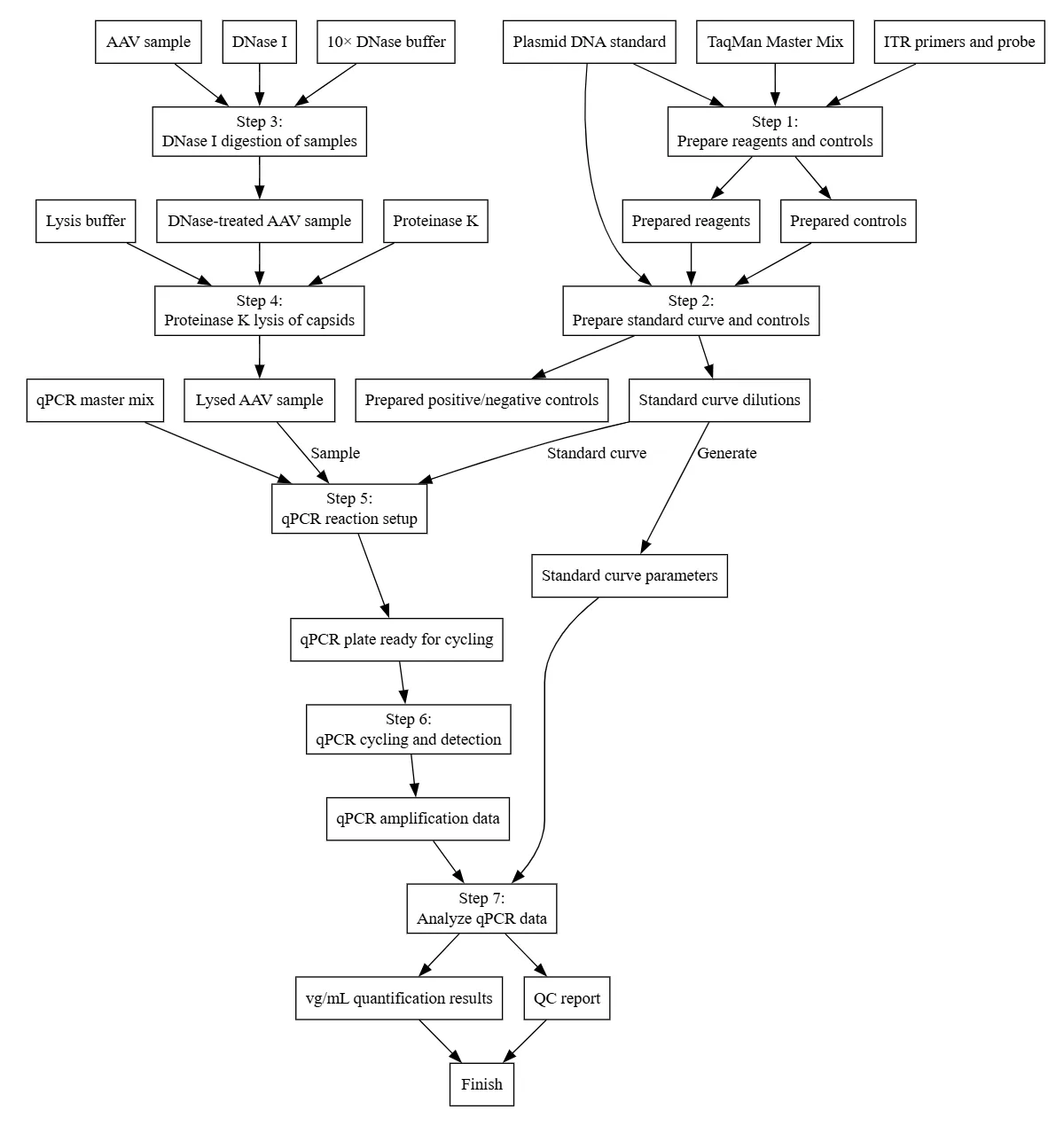
- Literature Review with Crossgrid: A structured comparison of papers on specificity, sensitivity, pretreatment, and acceptance criteria—each linked back to its source.
- Wet-Lab Protocol: Detailed SOP with reagents, plate map, master-mix prep, triplicate layout, and system suitability gates.
- Opentrons OT-2 Script: Automated plate setup with standard curves, metadata tracking, and user prompts for hand-offs.
Results
Tater produced a complete workflow in a single session with included regulatory requirements:
- Protocol with Evidence Across Literature: Justification for ITR-targeted TaqMan plus DNase and proteinase K based on comparative literature review.
- Embedded Acceptance Criteria: Clear thresholds for R², efficiency, triplicate CV, LOD/LOQ, and inhibition recovery.
- Regulatory-Aligned QC: Built-in criteria for specificity, accuracy, precision, linearity, range, LOD/LOQ, and robustness, per ICH Q2(R2).
- Reporting Schema: Templates for raw Ct tables, amplification plots, fitted standard curves, QC metrics, and calculated vg/mL with pass/fail flags.
Impact
By unifying literature review, wet-lab protocol design, regulatory expectations, and automation scripting into one deliverable, Tater transformed a multi-role, multi-week build into a reproducible workflow available on demand. This reduces time to data, increases reproducibility across operators and sites, and embeds regulatory confidence from the outset. The approach accelerates preclinical development while remaining extensible to clinical release testing, offering teams both immediate utility and a path to long-term compliance. In effect, Tater allows organizations to move from assay conception to automation-ready execution in hours rather than weeks, reshaping how critical assays are developed and validated.
| Workflow Stage | Manual (Typical Lab) | With Tater | Δ Improvement |
|---|---|---|---|
| Literature review & method comparison | 5 - 14 days | 13 min | ≈1,550× faster |
| Protocol drafting & MIQE validation | 6 - 10 hours | 8 min | ≈75× faster |
| Translating protocol to script | 3 weeks - 3 months | 55 min | ≈2,350× faster |
| Total end-to-end design cycle | 1 - 4 months | 1 hr 39 min | ≈1,750× faster |
Ready to try it yourself? Get started for free with our open access plan.
Citations
- AAV Titration by qPCR Using SYBR Green Technology V.2. https://dx.doi.org/10.17504/protocols.io.bawrifd6
- Sylvestre J, Conti-Permanne P. Emerging technologies & companies in cell & gene therapy manufacturing. Cell & Gene Therapy Insights 2022; 8(11), 1601–1649. https://dx.doi.org/10.18609/cgti.2022.232
- Liu Q, Grochowski CM, Bi W, Lupski JR, Stankiewicz P. Quantitative Assessment of Parental Somatic Mosaicism for Copy-Number Variant (CNV) Deletions. Curr Protoc Hum Genet. 2020 Jun;106(1):e99. https://dx.doi.org/10.1002/cphg.99
- Maturana CJ, Verpeut JL, Engel EA. Single-Cell Quantification of Triple-AAV Vector Genomes Coexpressed in Neurons. Curr Protoc. 2022 May;2(5):e430. https://dx.doi.org/10.1002/cpz1.430 . Erratum in: Curr Protoc. 2022 Jun;2(6): https://dx.doi.org/10.1002/cpz1.499
- Mazaika E, Homsy J. Digital Droplet PCR: CNV Analysis and Other Applications. Curr Protoc Hum Genet. 2014 Jul 14;82:7.24.1-7.24.13. https://dx.doi.org/10.1002/0471142905.hg0724s82