Engineering a Brighter GFP
Challenge
Designing new experiments typically requires days or weeks of manual effort—reviewing literature, designing constructs, enumerating variants, drafting protocols, and preparing analysis pipelines. Even for a simple idea like “make GFP brighter,” this process can be slow, fragmented, and error-prone. Researchers spend their time on protocol design, not scientific discovery.
Solution
We asked Tater, Potato's AI co-scientist, to help with a simple prompt:
I want to make a brighter GFP
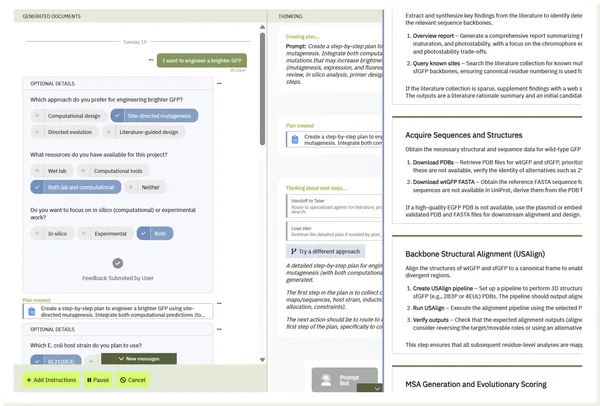
From this single request, Tater:
- Generated a curated literature review of brightness-enhancing mutations.
- Constructed a codon-optimized sfGFP scaffold sequence for E. coli.
- Designed a library of variants with both classical (S65T, F64L) and stabilizing superfolder substitutions.
- Produced a ready-to-run experimental protocol for cloning, expression, and OD600-normalized fluorescence screening.
- Delivered a structured dataset and documentation templates to standardize downstream analysis.
Results
Within minutes, Tater delivered a complete computational framework for engineering brighter GFP variants:
- Reference sequences built: Verified avGFP scaffold (UniProt P42212) and generated an sfGFP baseline.
- Variant library enumerated: Codon-optimized set including WT, F64L, S65T, Y145F, N149K, and A206K.
- Analysis plan prepared: Defined per-cell normalization (RFU/OD600), blank subtraction, and benchmarking controls.
- Protocol drafted: High-level plan for cloning into inducible vectors, expression in microplates, and fluorescence readout.
The result was a reproducible, literature-grounded workflow—ready for the lab without requiring hours of manual design.

Impact
This case study illustrates how Potato's AI agent compresses weeks of design work into a single interactive session. Instead of manually curating mutations, aligning references, and drafting protocols, scientists receive an end-to-end experimental plan they can immediately execute and refine.
By combining reasoning across literature with procedural design and data structuring, Tater enables researchers to move faster from idea → experiment → insight.
Ready to try it yourself? Get started for free with our open access plan.
Download Case Study as PDFCitations
- T.J. Barnard, X. Yu, N. Noinaj, J.W. Taraska, Crystal Structure of Green Fluorescent Protein (2014) https://doi.org/10.2210/pdb4KW4/pdb
- D. Sehnal, S. Bittrich, M. Deshpande, R. Svobodová, K. Berka, V. Bazgier, S. Velankar, S.K. Burley, J. Koča, A.S. Rose, Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures (2021) Nucleic Acids Research 49:W431-W437 https://doi.org/10.1093/nar/gkab314