Predicting Resistance in SARS-CoV-2 Main Protease
Challenge
Antiviral development is often hampered by resistance: viruses mutate, and drugs lose effectiveness. For SARS-CoV and SARS-CoV-2, the main protease (Mpro) is a key drug target—but testing every possible mutation in the lab is slow and expensive. Researchers need a way to quickly identify which single-nucleotide variants (SNVs) are likely to arise and remain functional, especially around inhibitor-binding sites.
Solution
We asked Tater, Potato's AI co-scientist, to:
“Compute evolutionary scores for all single-nucleotide missense variants in SARS-CoV main protease, and highlight the conservative substitutions near inhibitor-binding pockets.”
In response, Tater:
- Generated the complete set of 2,025 missense variants for Mpro (306 amino acids).
- Applied Jones–Taylor–Thornton (JTT) evolutionary scores to rank which substitutions are most likely to be tolerated.
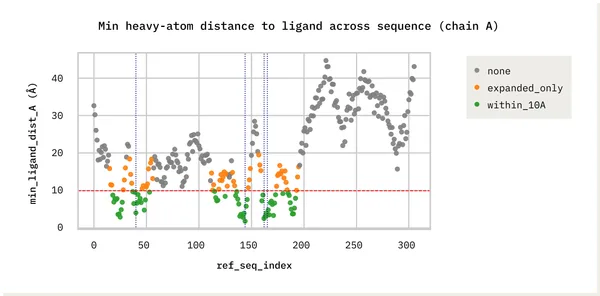
- Integrated multiple inhibitor-bound crystal structures to map variants near drug-binding sites.
- Produced a prioritized list of candidate mutations that could alter inhibitor sensitivity.
Results
Tater provided a biologically-grounded assessment of the resistance landscape to guide downstream functional testing:
- Variant scoring: Third-codon position changes were most tolerated, second-position least.
- Catalytic core: Critical residues like His41, Cys145, and Gly143 showed no tolerated single-base changes—confirming strong evolutionary constraints.
- Binding pockets: Prioritized conservative variants clustered around pocket residues such as Met49, His163, Glu166, and Met165, highlighting realistic hotspots for resistance.
- Outputs: Complete datasets, JTT score heatmaps, codon-position tolerance plots, and a top-25 ranked mutation set.
Impact
What normally requires a week of coding, structural analysis, and literature review was compressed into a single interactive session. The output gave researchers a ready-to-use dataset and prioritized mutation list to guide mutational scanning and clinical variant triage.
By combining evolutionary models with structural mapping, Tater makes it possible to anticipate resistance pathways before they emerge in patients—helping drug developers design more robust antivirals from the start.
Ready to try it yourself? Get started for free with our open access plan.
Download Case Study as PDFCitations
- Jones, D.T., Taylor W.R., Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 8:275-82 (1992). https://doi.org/10.1093/bioinformatics/8.3.275
- Jin, Z., Du, X., Xu, Y. et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582:289-93 (2020). https://doi.org/10.1038/s41586-020-2223-y
- Kneller, D.W., Li, H., Phillips, G. et al. Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease. Nat Commun 13:2268 (2022). https://doi.org/10.1038/s41467-022-29915-z
- Zhao, Y., Fang, C., Zhang, Q., et al. Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332. Protein Cell 13:689-693 (2022). https://doi.org/10.1007/s13238-021-00883-2